Explore Microbial Associations & Metabolic Interactions
MicrobiomeNet is a comprehensive database that brings together known statistical associations and genome-scale metabolic models (GEMs) to facilitate hypothesis generation and mechanistic insights. GEMs depict organisms' nutritional requirements which can be used to infer their potential metabolic interactions for their observed co-occurrence patterns. Please refer to our publication for more details. Some common tasks you can perform with MicrobiomeNet:
- Search a microbe of interest for its known co-occurrence patterns based on literature → Association Table & Network.
- Find out the overall metabolic & taxonomic distances, competition & complementarity profiles → Neighbourhood Map & Metabolism Compatibility Map.
- Examine detailed metabolic dependencies (global metabolism, pathways, reactions, enzymes) between microbial taxa → GEM & Pathway Network Visual Analytics.
Search Database
Please use OmicsForum for troubleshooting and discussions so the whole community can benefit (invitation code: xialab_omics!).
|
|
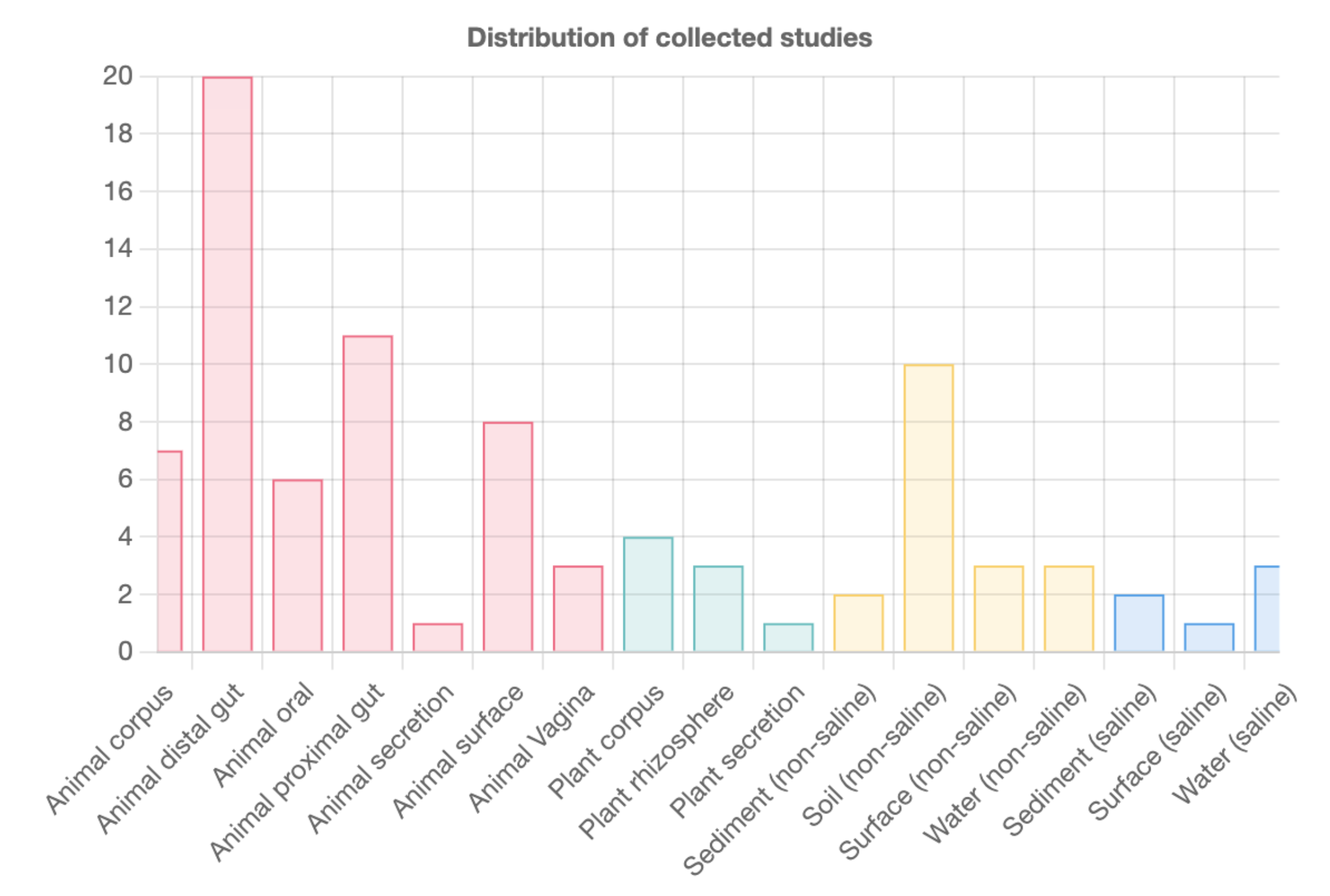
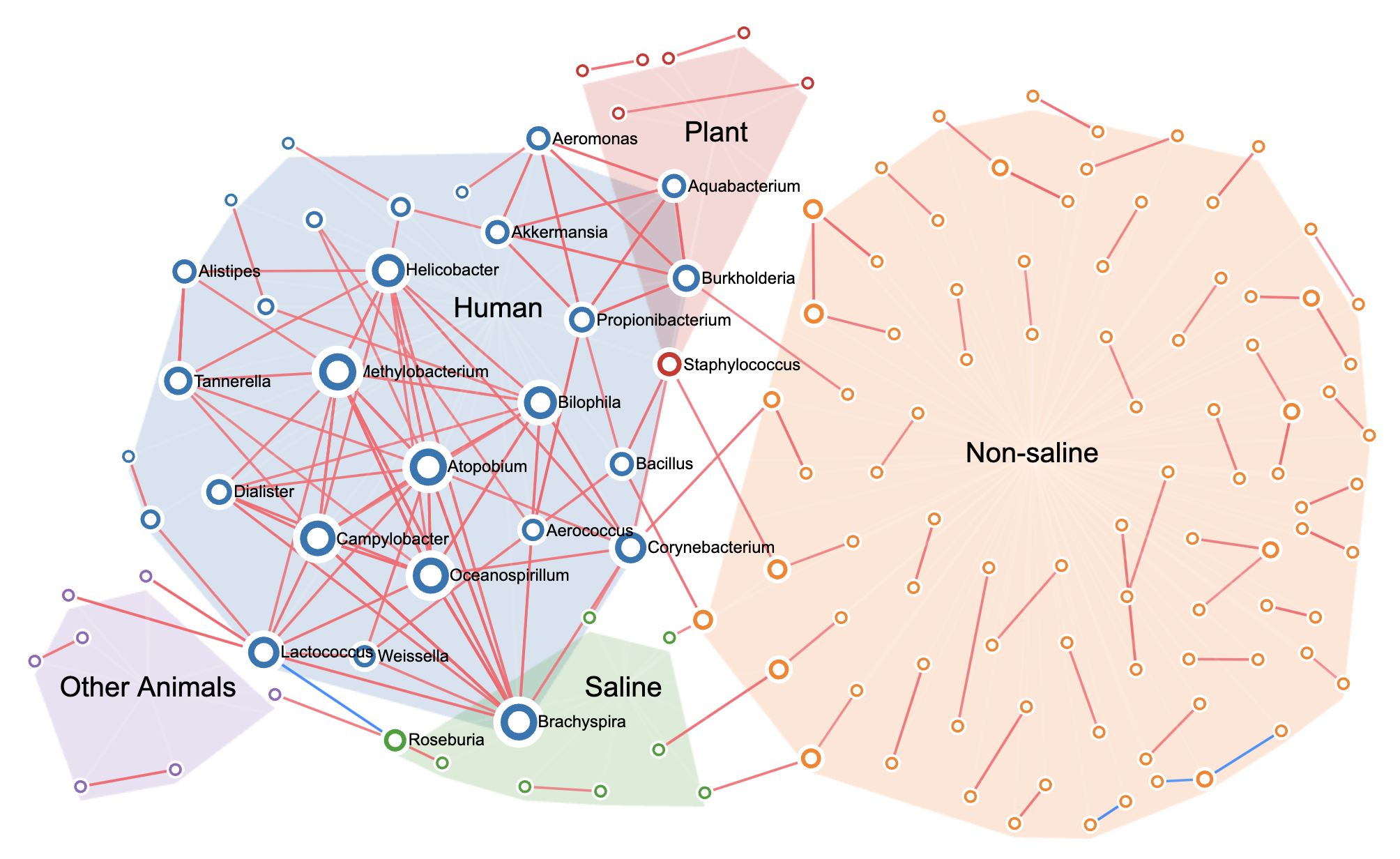
Microbial Association NetworksMicrobiomeNet currently contains ~ 6 million microbial statistical associations detected using different algorithms including Spearman, Pearson, SparCC, FlashWeave, MINE, Dice Index, and Binding. They have been collected from 76 studies covering diverse environments. Users can intuitively search and visually explore these networks. |
||||
|
||||
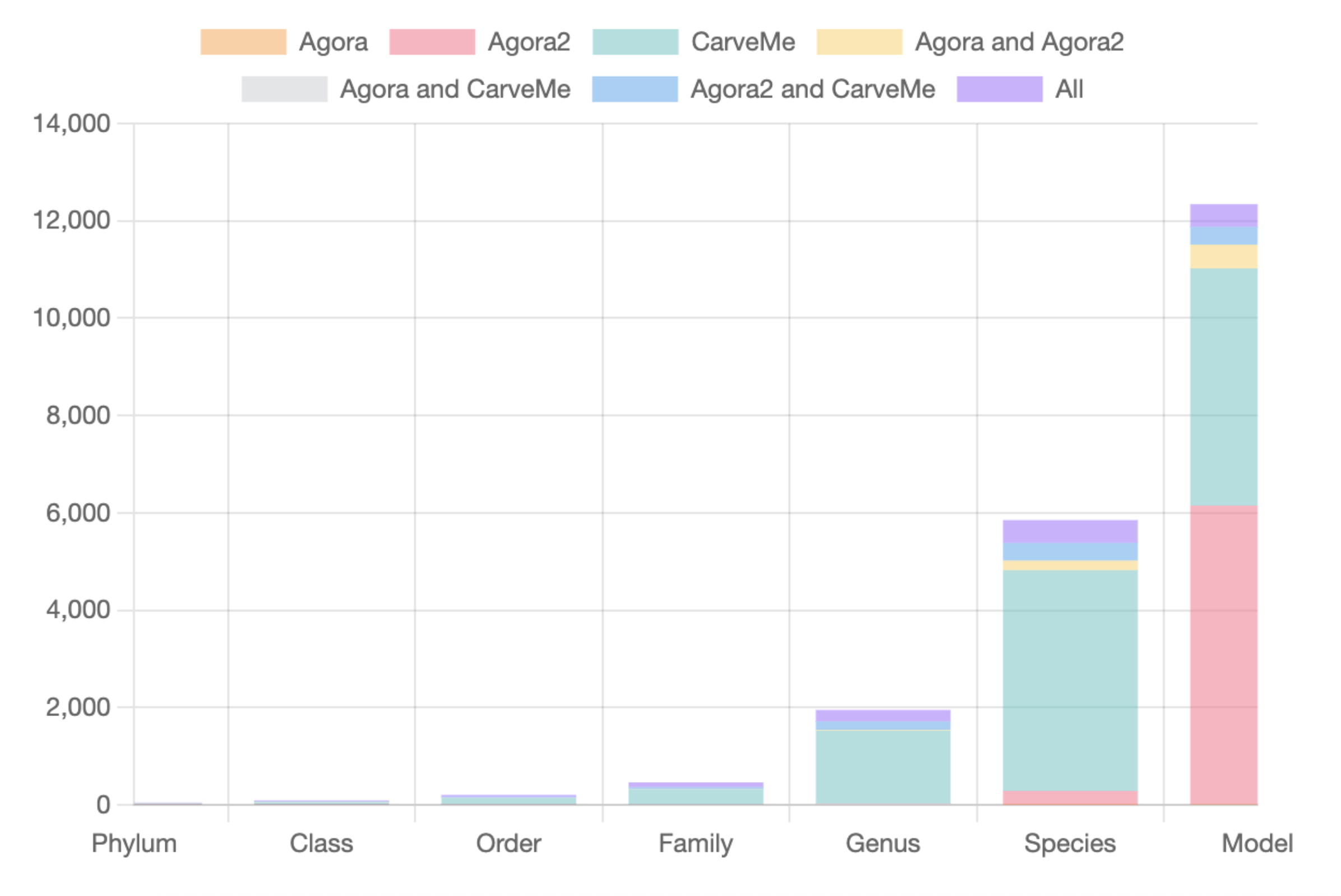
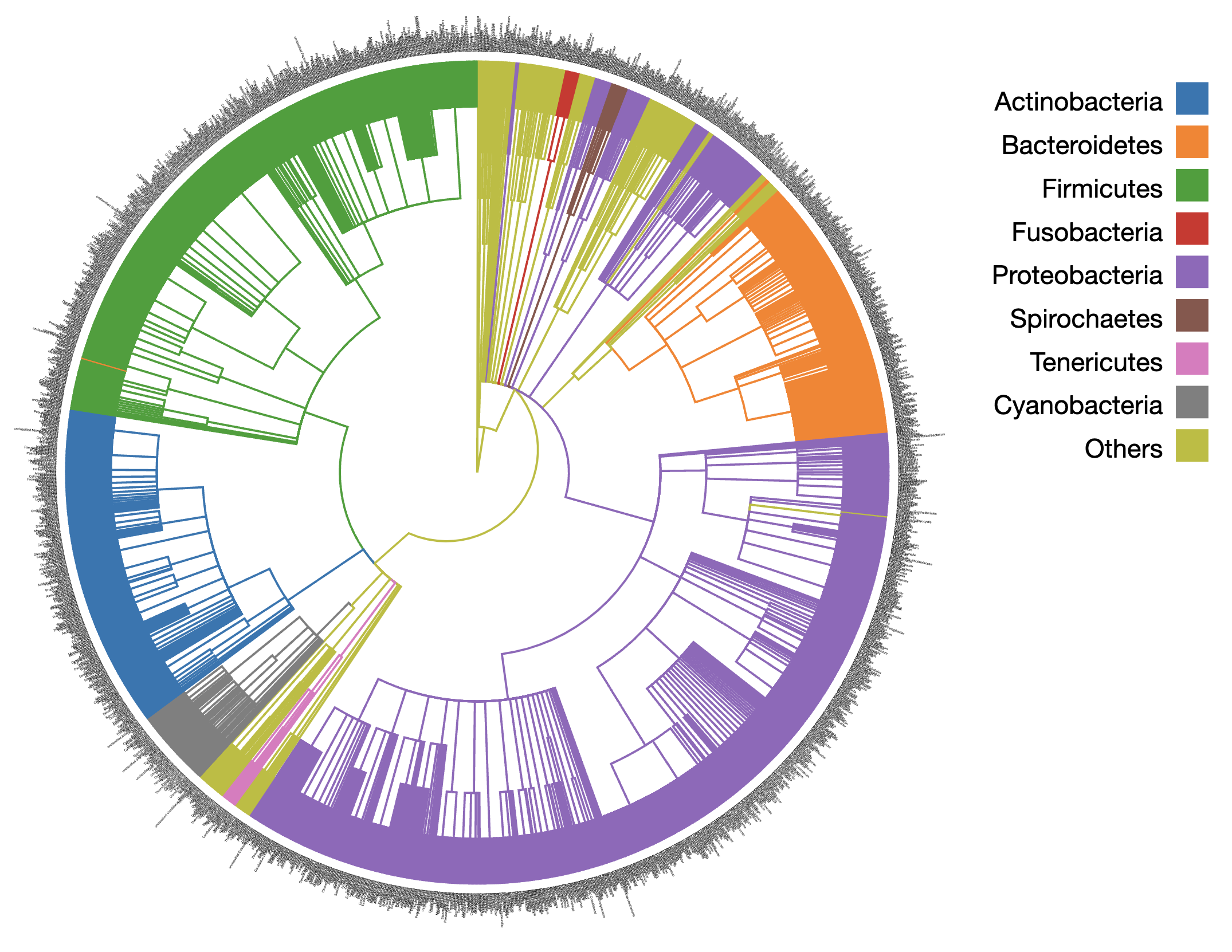
Genome-Scale Metabolic Models (GEMs)GEMs contain all known metabolic information including the genes, enzymes, reactions, and metabolites of a biological system. They are powerful tools to predict cellular metabolism and physiological states in living organisms. MicrobiomeNet currently contains 12,400 GEMs covering a broad ranges of taxonomies. Users can search, view and compare GEMs for microbes of interest. Pathways (also known as subsystems) are a set of reactions that share a similar metabolic function. MicrobiomeNet currently contains a total of 137 pathways extracted from the GEMs. |
||||
|
||||